Paradoxurus hermaphroditus (Pallas 1777)
Table of Contents
 |
| Baby common palm civet on a tree (Photograph by Chan Kwokwai[45]: Permission given.) |
I. Introduction
The common palm civet (Paradoxurus hermaphroditus) is fondly known as Musang in Malay and Toddy cat in English for its love of the sap used to make the alcoholic drink “toddy” 1. Though often labelled as the 'civet cat', the common palm civet is actually distantly related to cats and is instead more closely related to mongoose and hyenas 2, 3.
Its scientific name, Paradoxurus hermaphroditus, translates to paradoxical and hermaphrodite respectively. Coined in by Frédéric Curvier in 1821, "Paradoxurus" actually refers to the paradoxical tail of the civet, of which was thought to act like a fifth limb and looked very unlike the tails of its close relatives (hyenas and mongoose) 2, 5, 6. The species epithet "hermaphroditus", on the other hand, was named after hermaphrodite, a greek god who had both female and male sexual organs due to the resemblance of the civets’ scent glands to testicles and much confusion over identifying the sex of the civet. 1, 2
 |
| Common palm civet lazing on a branch with full view of tail and exposed bottom (Photograph by Nick Baker[39]: Permission given) |
The common palm civet is also a nocturnal creature and is thus largely active from dusk (1800h) to dawn (0400h)14, 15. Hence, most pictures of the civets were taken at night.
As one of the last remaining small mammals left in Singapore, the survival of the common palm civet over the years could probably be attributed to its ability to adapt and live around humans 4. Unfortunately, venturing out of forests and into the close proximity of humans has its consequences. The common palm civet has been facing numerous threats ranging from human-civet conflicts arising from interactions between civets and residents, its association to SARS and the unsustainable demand for kopi luwak.
II. Recognising a common palm civet
Identification of the common palm civet is mainly through its general appearances. However, if you do have the opportunity to take a closer look at it, it has other distinguishing characteristics such as its feet and teeth as well.
General appearances2, 7, 8
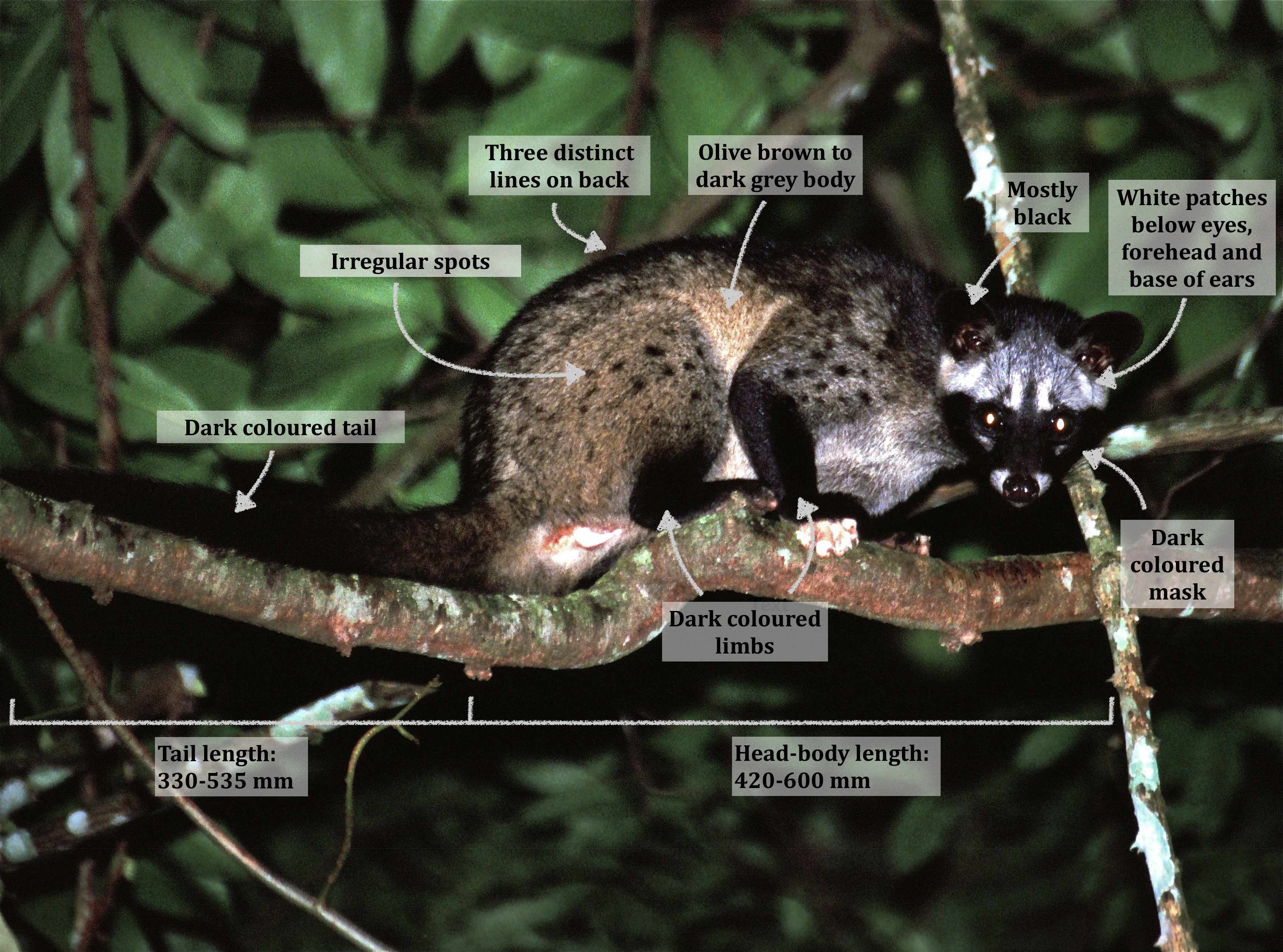 |
| General appearance of the common palm civet (Modified from photograph by Bernard Dupont[42]: CC-BY-SA 2,0) |
Just like how every individual is unique, some variation in outer appearance is expected to occur between individuals. For instance, the tail ends of some individuals might be white and the body colouration may differ from individual to individual. Nevertheless, the labelled diagram (above) shows the basic features of the common palm civet
Other distinguishing characteristics
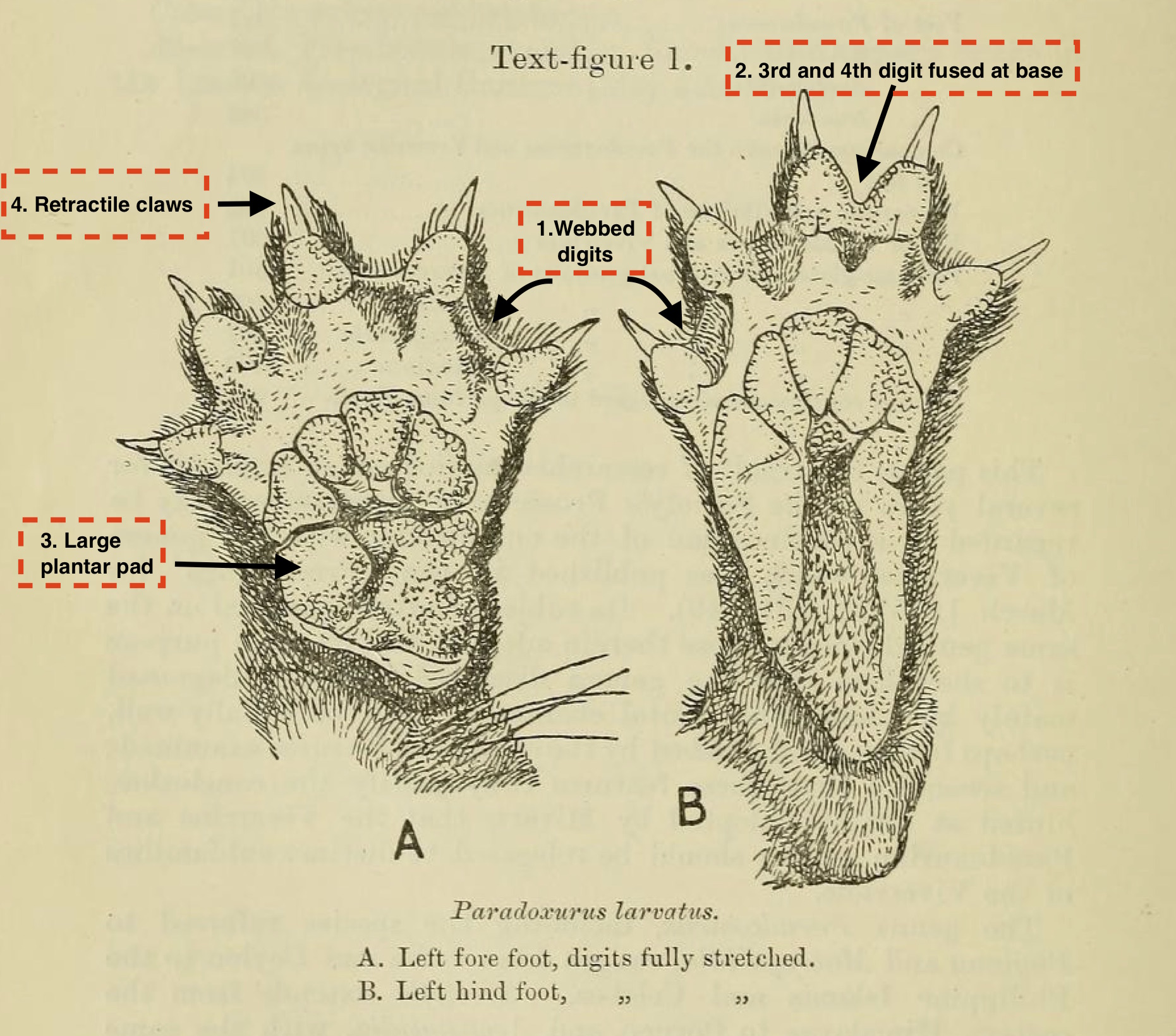 |
#1: Feet The common palm civet has plantigrade hind feet (B), which means it primarily walks with its feet planted on the ground like humans9. Distinct features of its feet includes:
These characteristics are also shared with other species within its genus10 such as Paradoxurus larvatus. For a more detailed description, have a look at this paper by Pocock et al. from the Biodiversity Heritage Library. |
||
|
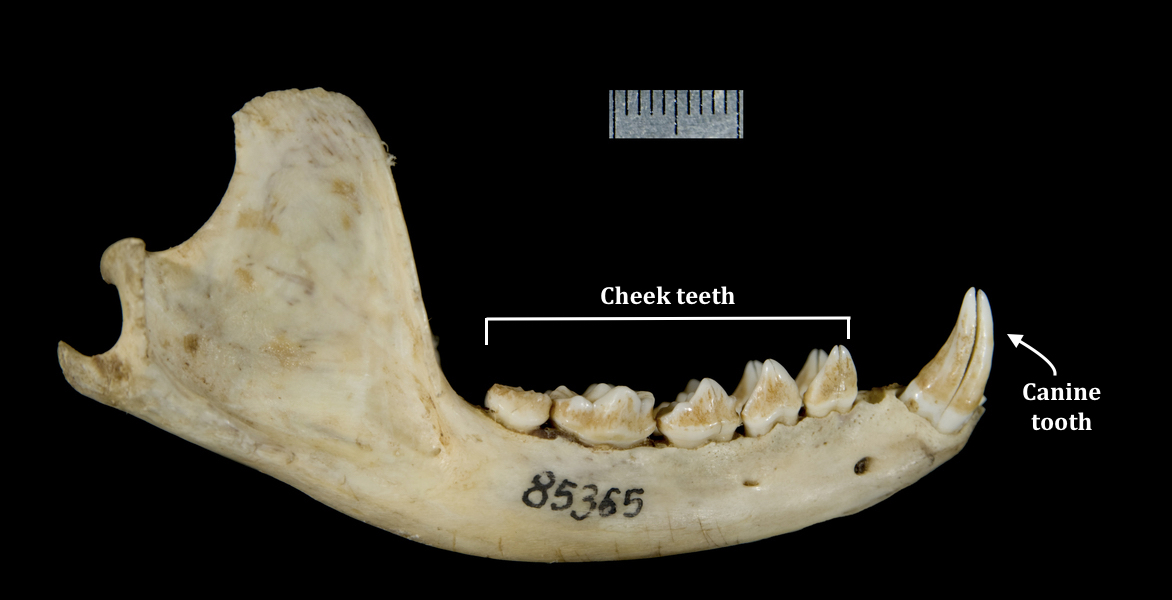
#2: Teeth The teeth of the common palm civet have developed over time to fit a broader diet. Hence you might notice that in comparison to other carnivores, the common palm civet has:
|
III. Civets of Singapore
The common palm civet is part of a larger group of civets.
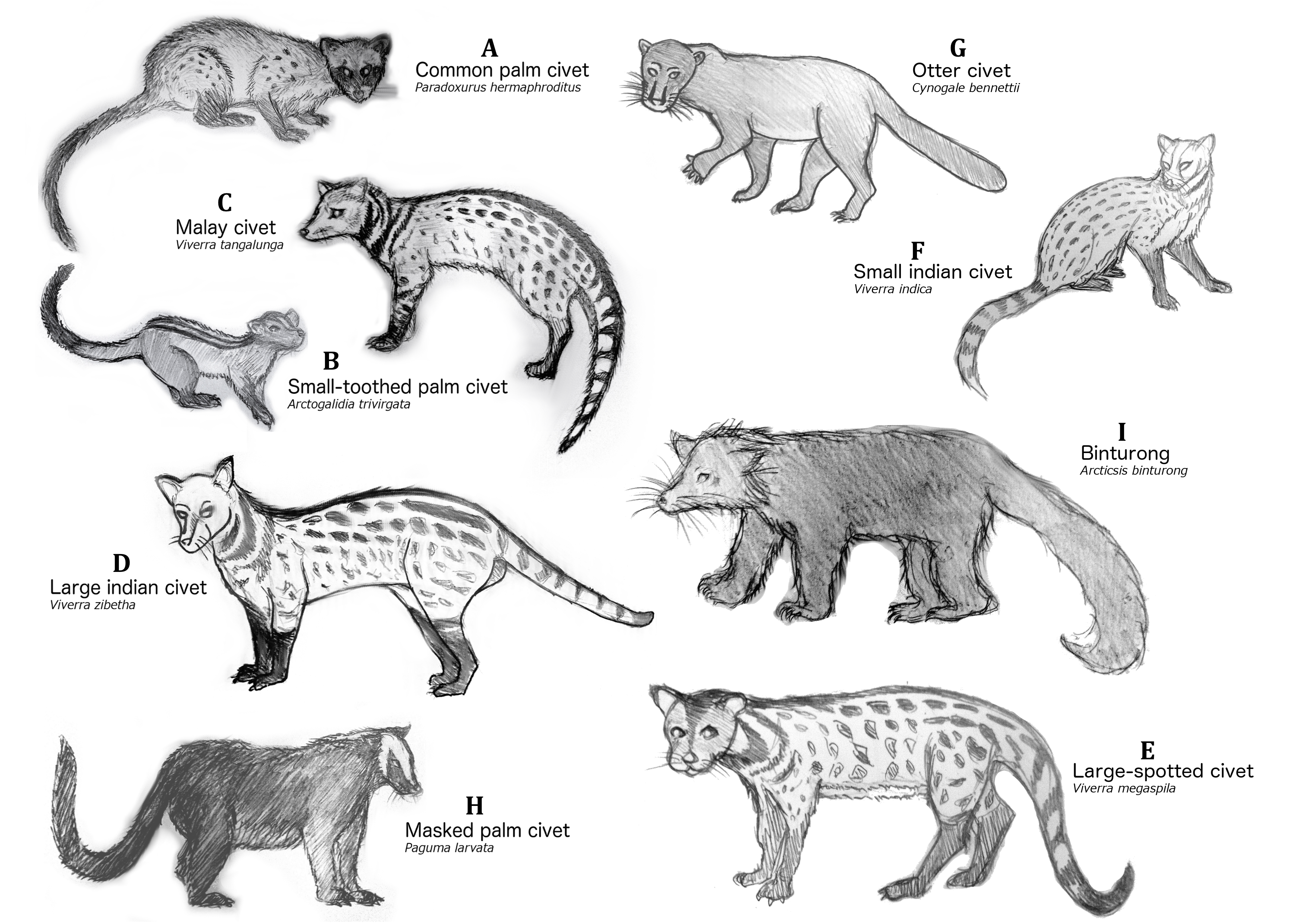 |
| Re-drawing of civets extinct and extant in Singapore by Alicia Ng, adapted from plates illustrated by Toni Ilobert, Medway7 and photographs of Bernard Dupoint42. |
To tell them apart, have a go at the flow chart below.
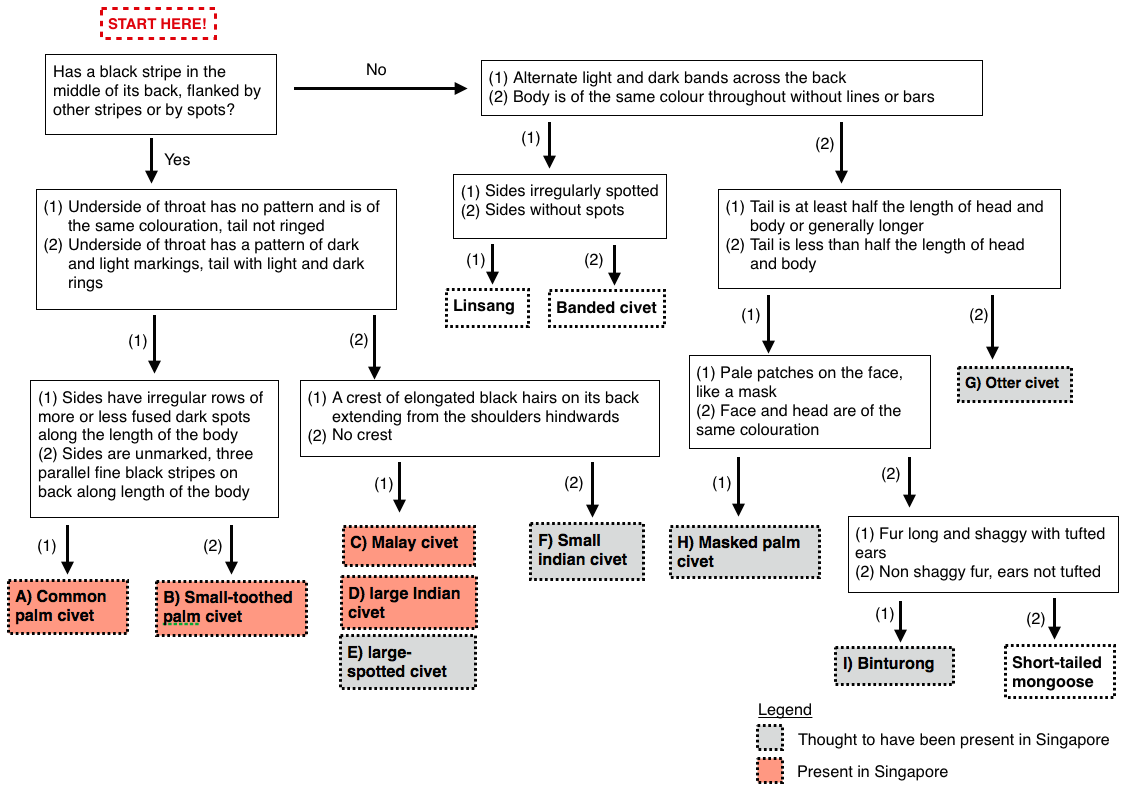 |
| Flow chart created adapted from key7 |
Out of the twelve possible species of civets in the flow chart, only four can be found in Singapore: the common palm civet, the Malay civet, the large indian civet and the small-toothed palm civet4.
|
Paradoxurus hermaphroditus Common name: Common palm civet Size: Medium Native: Yes Distinguishing features7, 8:
|
||
|
Viverra tangalunga Common name: Malay civet Size: Medium Native: Yes Distinguishing features7, 8:
|
||
|
Viverra zibetha Common name: Large indian civet Size: Medium Native: Yes Distinguishing features7, 8:
|
||
|
Arctogalidia trivirgata Common name: Small-toothed palm civet Size: Small Native: Yes Distinguishing features7, 8:
|
IV. Distribution
Local
Sightings of the four civets found in Singapore are shown in the map below 4. If you live near the Siglap or Opera estates, there might be a high chance you will encounter the common palm civet as there is a healthy population around that area 12.
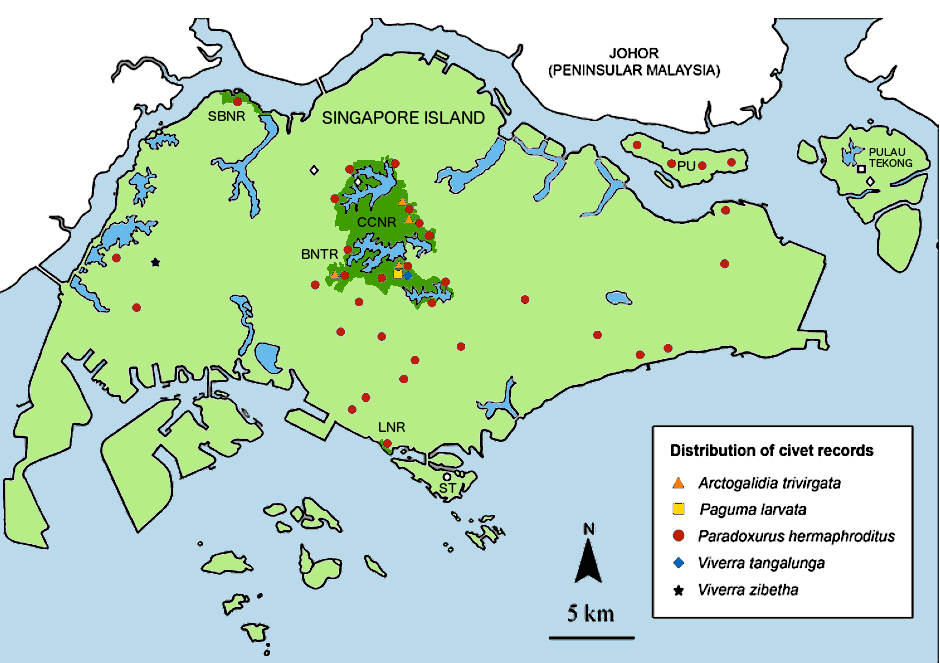 |
| Map of Singapore showing distribution of civets. Solid coloured shapes represent civet records. Hollow shapes are unconfirmed sightings. PU: Pulau Ubin, BTNR: Bukit Timah Nature Reserve, LNR: Labrador Nature Reserve, SBNR: Sungei Buloh Wetland Reserve, CCNR: Central Catchment Nature Reserve. (Modified from Chua et al., 2012 [4]) |
If you do see a civet, please take the time to fill in this form: http://mammal.sivasothi.com and help in the efforts to track our local fauna.
Global
On a larger scale, the common palm civet occupies most of Southeast Asia. Its geographic range extends from India in the West to the Philippines in the east, the Himalayas in the North and the islands of Indonesia in the South 8, 13. The map below shows the geographic range of Paradoxurus hermaphroditus13.
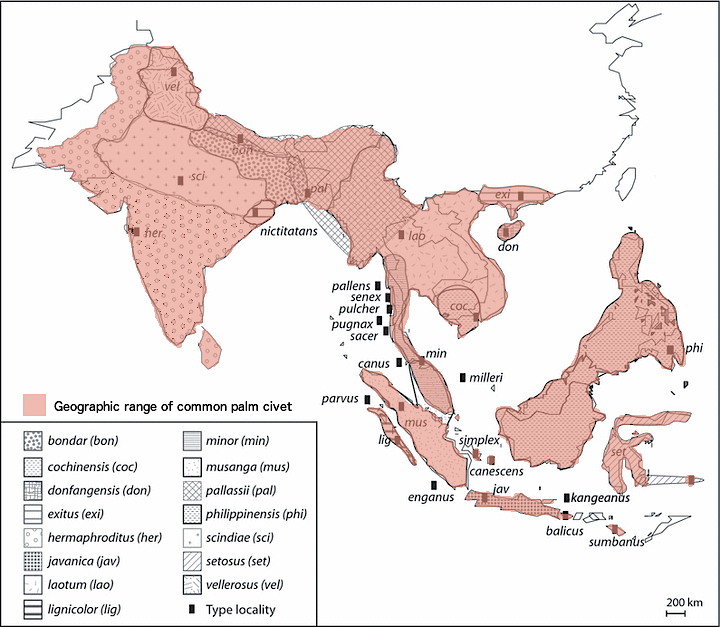 |
| Map of geographic range of Paradoxurus hermaphroditus (Modified from Patou et al.13) |
V. Habitat
The common palm civet has been known to inhabit a wide range of habitats, including mixed deciduous forest, shrub lands, tall secondary forests, plantations and gardens8, 14. During the day, the common palm civet may be found resting in tree cavities and in areas of dense vegetation15.
|
|
| Gif from video of common palm civet living in a tree hollow with her baby at Bidadari Cemetery (Video by 13seaeagle 48, created using GIPHY) |
VI. Behaviour
Though it spends most of its time up in the trees, the common palm civet do have the flexibility to head to the ground when crossing open spaces. They have also cleverly made use of roofs to head from one location to another7, 12.
It is also mostly solitary, but may form small groups consisting of the mother and its young1.
 |
| Civet family consisting of the mother and her young. (Photograph by Chan Kwokwai [47]: Permission given) |
The females are reputed to be territorial, while the males' territories may overlap2. To demarcate their territories, they largely communicate through scent markings left through a variety of methods18:
- Dragging of perineal gland (located at its belly) against the surface
- Rubbing of ear-neck area for objects above surface
- Heel rubbing on walking surface, where specific skin glands can release scent secretions
- Anal drag, often done after defecation
- Urination
- Defecation
Through just scent markings alone, the palm civet is able to discern the sex, species and familiarity of the scent’s owner.18
Curiously, civets have been increasingly known to use highways or routes as scent-marking locations despite the danger they face in becoming roadkill19. The common palm civet, in particular, uses gravel roads and roadside forests as a communication site through defecation. One of the reasons speculated for this strange habit was the presence of the civet’s food source, Ficus spp., along roadside forests, which may have led to its frequent visits to the area.
 |
| Civet faeces on road (Photograph by Jacqueline Chua [50]: Permission given) |
VII. Diet
The common palm civet is omnivorous and eats a mixed diet of fruits, leaves, insects, molluscs, small reptiles, birds and small mammals8, 15, 16. Their stomachs are well adapted to their diet and are highly tolerant of plant toxins that would have irritated our stomachs16. They also have teeth modified for a frugivorous diet2. As a result of their diverse diet, the common palm civet may switch to other food sources when one type of food is scarce14, 15. Fruits, however, still make up a huge proportion of their diet. Common palm civets are known to eat more than 10 species of fruits17. Amazingly, these seeds are generally undamaged after passing through the civet’s digestive system and are still able to germinate16, 17.
 |
| Civet faeces on Petai Trail at MacRitchie containing pulp of Tembusu fruits (Photograph by Jacqueline Chua 49: Permission given) |
VIII. Role in ecosystem
Due to its various attributes, the common palm civet is an excellent seed disperser. It helps to disperse the seeds of a variety of plant species ranging from fig trees (Ficus spp.), to cash crops like taro (Colocasia esculenta var. esculenta) to economically valuable timber trees such as Artocarpus spp.17, 19, 20.
For some seeds, such as the sugar palm, passing through the digestive gut of the civet is a requirement for germination16.
Besides persisting in disturbed habitats, the common palm civet is able to travel well over several hundreds of metres exceeding that of other disturbance tolerant animals such as rats and squirrels21. Coupled with their ability to travel between secondary and primary forests, the common palm civet could potentially widen the range of seed dispersal.
Moreover, given the differences in fruit preferences and seed handling, the common palm civet fulfils a unique role from other seed dispersers. For example, unlike other frugivores such as the hornbill and the spider monkey who are largely confined to trees, civets have been known to deposit seeds within the canopy, on the ground and in open areas such as stream banks and tree fall gaps, where conditions are ideal for germination37.
The common palm civet may hence play a vital role in seed dispersal and restoration of degraded forests in Singapore.
IX. Threats and conservation issues
 |
| Headshot of common palm civet (Photograph by Nick Baker [46]; Permission given) |
Although the common palm civet is noted as “least concern” under the IUCN red list and is not listed under the Singapore Red Data Book, its population status is poorly studied22. We do know however that the common palm civet is facing threats from human-civet conflict, misconceptions of SARS and the kopi luwak trade.
Human-civet conflict
In Singapore, common palm civets were thought of as nuisances. Besides raiding fruits from the gardens of residents and leaving behind faeces, residents were also agitated by the noises made by the civets when travelling across the roofs23. Residents also feared that the civets would cause harm or transmit diseases (SARS) to humans24.
As such, some of the residents have taken to trapping the civets23, 24. Though the-the majority of the residents have handed the civets over to the Night Safari to be displaced, a minority has expressed a worrying amount of rage towards the animals24. For instance, a local resident had trapped a common palm civet for eating the fruits of his papaya tree and damaging the telephone wires, threatening to barbeque the civet25. Though it is illegal to trap and kill a wild animal under section 5(1) of the Wild Animal and Birds Act, section 6(1) of the same act allows a person to kill the wild animal if it was found to have damaged the owner's property38. It is not known what has happened to the trapped civet.
 |
| Civet trapped for eating the fruits of the owner's papaya tree and gnawing on the telephone wires (Photograph by STOMP51: Permission given) |
Additionally, citizens have been known to keep the civets as pets, but grossly mistreat them either by ignorance or negligence.
| Mistreated civet in "A common palm civet found in a cage at Hougang car workshop" (Video by Straits Times 52) |
SARS
Back in 2004, a research paper linking the SARS epidemic to palm civets was released26. Two out of four SARS patients were found to have been in close proximity with the civets. They were waitress and customer respectively in a restaurant that serves civet meat in the form of an expensive prestigious dish: dragon, tiger and phoenix soup27. The six palm civets present in the restaurant were found to have a SARS-associated virus, prompting the extermination of 10,000 civets in Guangdong, China.
Later studies, however, found that the palm civets were not the source of the SARS virus, but may be involved in the transmission of SARS33. Notably, SARS is transmitted through other animals such as badgers and raccoon dog as well. Hence, recklessly killing off civets seems like a irrational solution to prevent the transmission of SARS when the focus should be on understanding and preventing the onset of SARS or other infectious diseases27, 33 . Moreover, civets and some species of animals may serve as a buffer between viruses and humans through slowing down the rate of transmission. Hence, we should not be too quick to attach negative connotations on civets.
Kopi luwak
An even bigger threat to the common palm civet is the demand for kopi luwak. Kopi luwak is coffee brewed from beans that have passed through the digestive system of civets28. The coffee beans are remarked to be more fragrant and flavourful than normal coffee29.
Here is a short introduction to kopi luwak:
| An introduction to Kopi luwak: "Extreme Coffee -- Sumatra" (Video by Journeyman Pictures53) |
The origins of kopi luwak allegedly came about because workers were denied coffee beans from the plantation. Hence to get their dose of coffee, they collected, cleaned and roasted the coffee beans collected from the faeces of wild palm civets29.
Kopi luwak was never meant to be a commercially viable good given the tediousness and difficulty of harvesting the defecated seeds28. The novelty and popularity of kopi luwak however soon led to a drastic increase in prices. In Singapore, a cup of kopi luwak costs a whopping $2530.
It is of little surprise that the pressure to supply the goods ultimately translates for the need to mass produce kopi luwak. Most kopi luwak are thus rarely “wild-sourced” and are instead farmed29. The civets are captured from the wild and fed exclusively on a diet of coffee cherries, which tends to have detrimental effects on their health28, 29. As mentioned earlier, civets have a broad diet and though they may switch from source to source, they still require a variety of food. Feeding them caffeine ladened coffee cherries nutritionally starves them and cause great mental distress, so much so that some civets exhibit self-harm. Consequently, trapping the civets into small cages drives them to display stereotypical behaviour such as restless repetitive pacing as shown in the video below.
This video summarises the cruelties imposed on the civets due to our demand for more “fragrant coffee”:
| Video summarising the cruelties of the civet coffee industry (Video by PETA UK54) |
For more information on Kopi Luwak and to support of the ban on kopi luwak, please visit this site: https://projectluwaksg.wordpress.com!
X. Taxonomy and systematics
Original description
 |
| Original description of Paradoxurus hermaphroditus by Pallas 1777, accessible by Biodiversity Heritage Library 35 |
The common palm civet (Paradoxurus hermaphroditus) was first described by Peter Simin Pallas, a German botanist and zoologist, in 1777 under the binomial name: “Viverra hermaphroditus”. The original description was published in the book: Die Säugethiere in Abbildungen nach der Natur, mit Beschreibungen35. A rough translation of the text reveals a brief description of the various defining morphological features of the common palm civet, including its fur coloration and its relative proportions.
Taxonavigation
| Class |
Mammalia |
| Order |
Carnivora |
| Family |
Viverridae |
| Genus |
Paradoxurus |
| Species |
Paradoxurus hermaphroditus |
Systematics
Prior to the availability of molecular data, there were numerous hypothesis for the classification of taxa within the family of Viverridae31.For instance, Mivart’s classification found three main taxa based on characters such as the perineal scent glands, skulls, dentition, colouration and foot structure:
- Genetta, Viverricula, Viverra, Fossa, Prionodon and Poiana
- Paradoxurus, Arctogale, Hemigalea, Arctictis, Nandinia
- Cynogale
Whereas, Pocock in contrast has outlined seven taxa, basing his classification on detailed analysis of perineal scent glands:
- Viverrinae, Fossinae (True civets)
- Prionodontinae (Asian linsang)
- Paradoxurinae (The true palm civets)
- Arctogalidinae
- Hemigalinae (Banded civets)
- Cynogalinae (Otter civets)
- Nandiniidae (African palm civets)
Wovencraft (1984) reclassified the taxa to six subfamilies:
- Hemigalinae (Banded and otter civets)
- Paradouxurinae (The true palm civets)
- Viverrinae (True civets)
- Nandiniinae (African palm civets)
- Cryptoproctinae (Malagasy civets)
- Euplerinae (Malagsy viverrids)
- Hemigalinae (Banded and otter civets)
- Paradoxurinae (The true palm civets)
- Genettinae (Genets)
- Viverrinae. (True civets)
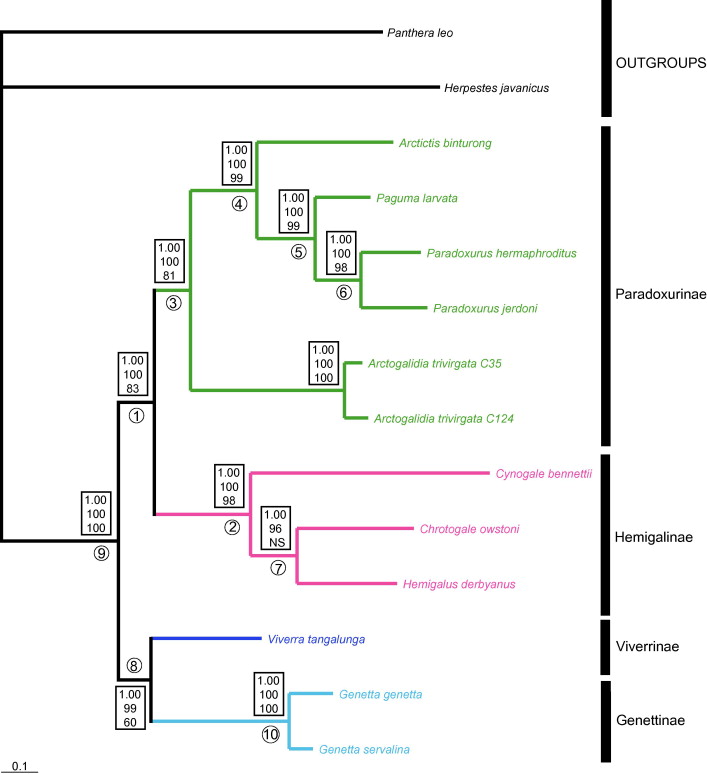
Phylogeny
Most of the taxonomic problems with regard to the common palm civet occurred at the higher taxa. Hence, phylogenetic analyses were conducted on the subfamilies of Viverrids (Family Viverridae) by Veron and Heard (2000)32, Gaubert and Cordeiro-Estrela (2006)3 and Patou et al. (2008)34, which revealed the relationships within subfamilies of civets and genets to be relatively consistent. With phylogenetic analyses using molecular data, the confusion regarding the arrangement of subfamilies within the Viverrids is proposed to contain only four subfamilies.
These are the phylogenetic trees from the various analyses and a brief description of the data and optimality criteria used for each tree:
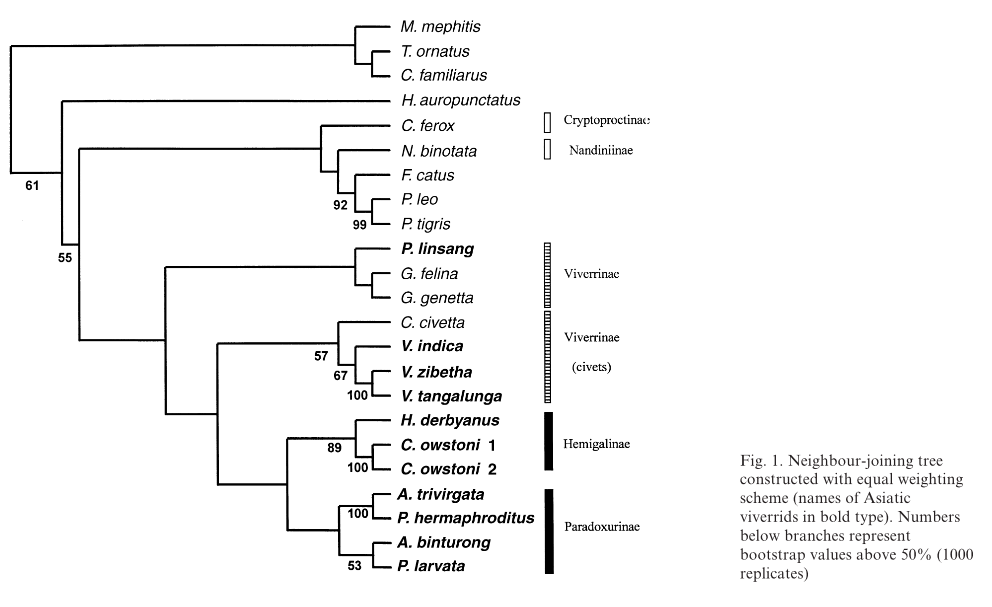
| Veron and Heard32 constructed the phylogenetic tree based on molecular data (a 790bp cytochrome b sequence, of which 664bp was used in analysis). Maximum Parsimony and Neighbour-joining were the optimality criteria for the tree. |
|
||||
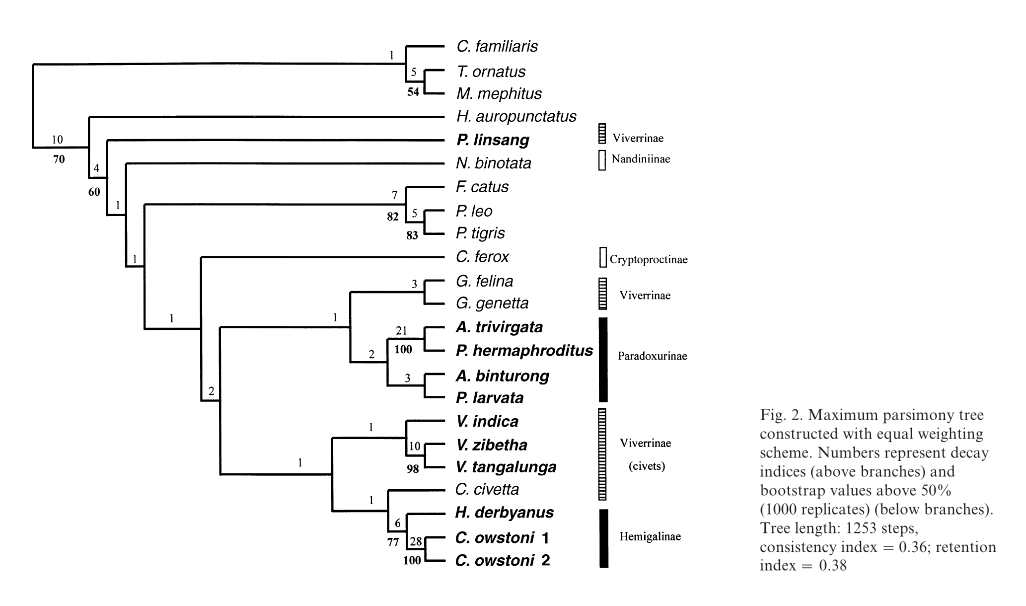
| The phylogenetic tree of Viverrinae by Gaubert and Cordeiro-Estrela3 utilises four sets of genes cytochrome b, transthyretin intron I and IRBP exon 1 (IRBP1) to generate a combined data set through Bayesian analysis. Optimality criteria Maximum Parsimony, Maximum Likelihood were also considered in the analysis. |
|
||||
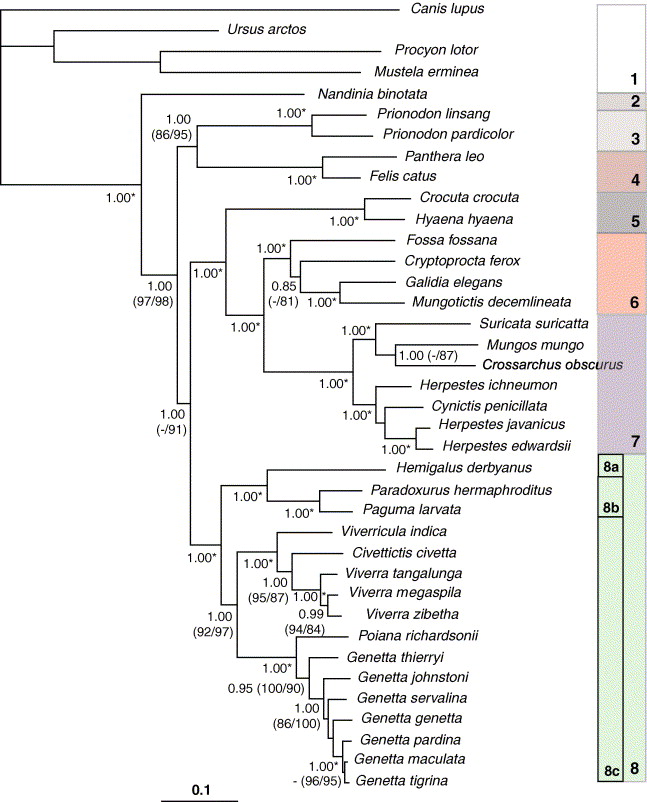
| Patou et al.34 derived its phylogenetic tree through the use of molecular data. Four genes were sequenced: two mitochondrial genes (Cytochrome b and ND2) and two nuclear genes (b-fibrinogen intron 7 and IRBP exon 1). Maximum Parsimony, Maximum Likelihood and Bayesian inference were used as optimality criteria. The datasets were then combined to reconstruct the final phylogenetic tree seen below. |
|
As seen from the phylogenetic trees, phylogenetic relationships between P. hermaphroditus and sister taxa were largely found to be supported and consistent. For instance, within Paradoxurinae, the phylogenetic relationships between (Arctictis binturong (Paguma larvata (P. hermaphroditus, P. jerdoni))) were found to be stable in all phylogenetic trees. In most of the phylogenetic trees, the clade (Hemigalinae, Paradoxurinae), comprising of Asian palm civets was shown to be a well-supported relationship34. With the exception of the phylogenetic tree analysed using Maximum Parsimony in Veron and Heard32, the (Genettinae+Viverrinae) was found to be the sister group of the Asian palm civets (Hemigalinae, Paradoxurinae).
Additionally, the phylogenetic studies by Patou et al.34 and Gaubert and Cordeiro-Estrela3, have tentatively concluded that the family Viverridae consists of four subfamilies: Genettinae, Viverrinae , Hemigalinae, and Paradoxurinae.
XI. Further resources
Other species pages
Local news of the common palm civet
Citizen science
Kopi luwak
XII. Literature cited
Literature
1.“Common palm civet (Paradoxurus hermaphroditus) at the Shores of Singapore,” by WildSingapore. 2013. URL: http://www.wildsingapore.com/wildfacts/vertebrates/mammals/hermaphroditus.htm
2.“A phylogenetic reappraisal of the Viverridae and its relationship to other carnivora (civets, mongooses, herpestidae).” by Wozencraft, W. C.. 1984. URL: http://search.proquest.com.libproxy1.nus.edu.sg/docview/303321405?pq-origsite=summon
3. Gaubert, P. & Cordeiro-Estrela, P., 2006. Phylogenetic systematics and tempo of evolution of the Viverrinae (Mammalia, Carnivora, Viverridae) within feliformians: implications for faunal exchanges between Asia and Africa. Mol. Phylogenet. Evol. 41, 266–78.
4. Chua, M. a. H., Lim, K. K. P. & Low, C. H. S., 2012. The diversity and status of the civets (Viverridae) of Singapore. Small Carniv. Conserv. 47, 1–10.
5. “Paradoxurus - Animalia.,” by Animalia. 2015. URL: http://metazoa.us/paradoxurus
6. “Paradoxure - definition, etymology and usage, examples and related words.,” by Fine Dictionary, Webster’s Revis. Unabridged Dict. 2015. URL: http://www.finedictionary.com/Paradoxure.html
7. “The wild mammals of Malaya (Penisular Malaysia) and Singapore Second Edition,” by Medway, Lord, 1978. Oxford Univesity Press.
8. “A Field Guide to the Mammals of South-East Asia.,” by Francis, C. M. & Barrett, P., 2008. New Holland Publishers. URL: https://books.google.com/books?id=zWeS8A6nunIC&pgis=1
9. Taylor, M. E., 1988. Foot structure and phylogeny in the Viverridae (Carnivora). J. Zool. 216, 131–139.
10. Pocock, R. I., S., F. R., S., F. L. & S., F. Z, 1915. On the feet and glands and other external characters of the Paradoxurine genera Paradoxurus, Arctictis, Arctogalidia, and Nandinia. Proc. Zool. Soc. London, 387–412.
11.“ADW: The Diversity of Cheek Teeth. he Anim. Divers. Web” by Myers, P., R. Espinosa, C. S. Parr, T. Jones, G. S. Hammond, and T. A. D., 2015. URL: http://animaldiversity.org/collections/mammal_anatomy/tooth_diversity.
12. “Musang facing threat from annoyed residents.,” by Ang, Y., 2010.
13. Patou, M.-L. et al., 2010. Evolutionary history of the Paradoxurus palm civets - a new model for Asian biogeography. J. Biogeogr. 37, 2077–2097.
14. Su, S. & Sale, J., 2007. Niche differentiation between Common Palm Civet Paradoxurus hermaphroditus and Small Indian Civet Viverricula indica in regenerating degraded forest, Myanmar. Small Carniv. Conserv. 36, 30–34.
15. Joshi, A. R., David Smith, J. L. & Cuthbert, F. J., 1995. Influence of Food Distribution and Predation Pressure on Spacing Behavior in Palm Civets. J. Mammal. 76, 1205–1212.
16. Bartels, E., 1964. On Paradoxurus hermaphroditus javanicus (Horsfield, 1824). The Common Palm Civet or Toddy Cat in western Java. Notes on its food and feeding habits. Its ecological importance for wood and rural biotopes. Beaufortia 10, 193–201.
17. Jothish, P. S., 2011. Diet of the Common Palm Civet Paradoxurus hermaphroditus in a rural habitat in Kerala, India, and its possible role in seed dispersal. Small Carniv. Conserv. 45, 14–17.
18. Rozhnov, V. V. & Rozhnov, Y. V., 2003. Roles of Different Types of Excretions in Mediated Communication by Scent Marks of the Common Palm Civet, Paradoxurus hermaphroditus Pallas, 1777 (Mammalia, Carnivora). Biol. Bull. 30, 584–590.
19. Nakabayashi, M., Nakashima, Y., Bernard, H. & Kohshima, S., 2014. Utilisation of gravel roads and roadside forests by the common palm civet (Paradoxurus hermaphroditus) in Sabah, Malaysia. Raffles Bull. Zool. 62, 379–388.
20. “The Dispersal of Taro by Common Palm Civets.,” by Hambali, G. G., 1979. in Int. Symp. trop root tuber Crop. Symp. Proc. URL:http://www.istrc.org/images/Documents/Symposiums/Fifth/5th_symposium_proceedings_0050_545.pdf
21. Nakashima, Y., Inoue, E., Inoue-Murayama, M. & Abd. Sukor, J., 2010. Functional uniqueness of a small carnivore as seed dispersal agents: a case study of the common palm civets in the Tabin Wildlife Reserve, Sabah, Malaysia. Oecologia 164, 721–730.
22.“Paradoxurus hermaphroditus (Common Palm Civet, Mentawai Palm Civet).,” by Duckworth, J. W. et al., 2008. IUCN. URL:http://www.iucnredlist.org/details/41693/0
23. Jaffar, R., Chandran, S. & Pillai, K., 2012. Living with common palm civets (Paradoxurus hermaphrodites): a civet conservation and education initiative by the Night Safari. in South East Asian Zoos Aquariums. URL:http://www.researchgate.net/publication/268149579_LIVING_WITH_COMMON_PALM_CIVETS_(Paradoxurus_hermaphrodites)_A_CIVET_CONSERVATION_AND_EDUCATION_INITIATIVE_BY_THE_NIGHT_SAFARI
24.“The great ‘musang’ stakeout.,” by Ang, Y., 2009. URL:http://wildsingaporenews.blogspot.sg/2009/11/great-musang-stakeout.html#.VkAlZISGVUR
25. “Bungalow owner wants to barbeque civet cat for destroying telephone wires.,” by STOMP, 2012. URL:http://singaporeseen.stomp.com.sg/this-urban-jungle/bungalow-owner-wants-to-barbeque-civet-cat-for-destroying-telephone-wires>
26. Wang, M. et al., 2005. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 11, 1860–5.
27.“SARS Scapegoat? China Slaughtering Civet Cats.,” byLovgren, S., 2003. Natl. Geogr. News (2003). URL:http://news.nationalgeographic.com/news/2004/01/0109_040109_SARS.html
28. “Civet coffee: why it’s time to cut the crap.” by Wild, T., 2013. The Guardian. URL:http://www.theguardian.com/lifeandstyle/wordofmouth/2013/sep/13/civet-coffee-cut-the-crap
29. “The World’s Most Expensive Coffee Is a Cruel Cynical Scam, ” by Kwok, Y., 2013. Time (2013). URL:http://world.time.com/2013/10/02/the-worlds-most-expensive-coffee-is-a-cruel-cynical-scam
30.“Where to find Luwak Coffee In Singapore?,” by William, 2013.URL:http://blog.omy.sg/safra/2013/11/29/where-to-find-luwak-coffee-in-singapore
31. Gregory, W. K. & Hellman, M., 1939. On the Evolution and Major Classification of the Civets (Viverridae) and Allied Fossil and Recent Carnivora: A Phylogenetic Study of the Skull and Dentition. Proc. Am. Philos. Soc. 81, 309–392.
32. Veron, G. & Heard, S., 2000. Molecular systematics of the Asiatic Viverridae (Carnivora) inferred from mitochondrial cytochrome b sequence analysis. J. Zool. Syst. Evol. Res. 38, 209–217.
33. Wang, L.-F., & Eaton, B. T. 2007. Bats, Civets and the Emergence of SARS. In J. E. Childs, J. S. Mackenzie, & J. A. Richt (Eds.), Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission (Vol. 315, pp. 332–334). Berlin, Heidelberg: Springer Berlin Heidelberg. doi:10.1007/978-3-540-70962-6_13
34. Patou, M.-L. et al., 2008. Phylogenetic relationships of the Asian palm civets (Hemigalinae & Paradoxurinae, Viverridae, Carnivora). Mol. Phylogenet. Evol. 47, 883–92.
35. Goldfuss, G. A., Schreber, J. C. D. & Wagner, A. J. Die Säugthiere in Abbildungen nach der Natur, mit Beschreibungen /. (Expedition des Schreber’schen säugthier- und des Esper'schen Schmetterlingswerkes ..., 1774). doi:10.5962/bhl.title.67399
36. “Carnivoran Evolution: New Views on Phylogeny, Form and Function.,” by Cambridge University Press, 2010. URL: https://books.google.com/books?id=HPw0C2i8QXkC&pgis=1
37. Chakravarthy, D., & Ratnam, J. 2015. Seed dispersal of Vitex glabrata and Prunus ceylanica by Civets (Viverridae) in Pakke Tiger Reserve, north-east India: spatial patterns and post-dispersal seed fates. Tropical Conservation Science, 8(2), 491.
38. Wild Animals and Birds Act, Rev. ed. Cap 351, s. 6(1) (2000)
Photographs and Videos
39. “Male relaxing on a thick liana at Sungei Relau, Taman Negara, Pahang, Peninsular Malaysia,” by Nick Baker. Ecology Asia. URL: http://www.ecologyasia.com/verts/mammals/common_palm_civet.htm
40. “Large Indian Civet, Viverra zibetha in Kaeng Krachan national park,” by Tontantravel. Flickr. 14 Nov 2014. URL:https://www.flickr.com/photos/tontantravel/15924839626/in/photolist-qgdY1s-rQM5XJ-6tMQoc-5b8jRD-n9Zyr4-bXRqko-bq9grE-bq9gLE
41. “Small-toothed Palm Civet, Arctogalidia trivirgata in Khao Yai national park,” by Tontantravel. Flickr. 30 Jul 2014. URL: https://www.flickr.com/photos/tontantravel/20757398043/
42. “Common Palm Civet (Paradoxurus hermaphroditus),” by Bernard Dupont. Flickr. Sep 2003. URL: https://www.flickr.com/photos/berniedup/7781509830/
43. “Malay civet,” by bluefuton. Flickr. 12 Sep 2008. URL: https://www.flickr.com/photos/bluefuton/2907140166/
44. “Lateral view of lower jaw,” by Phil Myers, Museum of Zoology, University of Michigan-Ann Arbor. Animal Diversity Web. 11 Dec 2008. URL: http://animaldiversity.org/accounts/Paradoxurus_hermaphroditus/specimens/collections/contributors/phil_myers/ADW_mammals/specimens/Carnivora/Viverridae/Paradoxurus_hermaphroditus/lower_lateral/
45. “Civet on a rain tree.,” by Chan Kwokwai. Flickr. 10 April 2011. URL:https://www.flickr.com/photos/kwokwai76/8061780809/in/album-72157631709562308/
46. “Young male, in a mango tree in the Portsdown area of Singapore,” by Nick Baker. Ecology Asia. URL: http://www.ecologyasia.com/verts/mammals/common_palm_civet.htm
47. “A Mum with 2 kittens” by Chan Kwokwai. Flickr. 10 April 2011. URL:http://wildpix.blogspot.sg/2011/04/common-palm-civet-of-siglap-estate.html
48. “Common/Asian palm civet n Baby. Former Bidadari Cemetary Singapore.,” by 13seaeagle. 9 October 2015. URL:https://www.youtube.com/watch?v=xQgDLbLbWGE
49. “Civet faeces on Petai Trail at MacRitchie containing pulp of Tembusu fruits,” by Jacqueline Chua.
50. “Civet faeces on road,” by Jacqueline Chua
51. “Bungalow owner wants to barbeque civet cat for destroying telephone wires,” by STOMP. 29 March 2012. URL: http://singaporeseen.stomp.com.sg/this-urban-jungle/bungalow-owner-wants-to-barbeque-civet-cat-for-destroying-telephone-wires
52. “A common palm civet found in a cage at Hougang car workshop,” by Straits Times. 6 May 2015. URL: https://www.youtube.com/watch?v=1Rp12P4sFx8
53. “Extreme Coffee -- Sumatra,” by Journeyman Pictures. 31 Oct 2007. URL: https://www.youtube.com/watch?v=wCG31fSAr4M
54. “Kopi Luwak: Cruelty in Every Cup,” by PETA UK. 17 Sep 2013. URL: https://www.youtube.com/watch?v=leSQt1NeCok